Bringing Data To Life - DIY
Source code and documentation to COVIDBlooms, my COVID-19 data visualisation.
Last week I presented and described the why behind a recent COVID-19 data visualisation exercise I undertook recently.
The primary purpose of that exercise was of course, to enable detailed data analysis on a relevant and prominent topic. But there was an ulterior motive too - these exercises never fail to teach things along the way. Staying current and fluent in data science tools is important to me, because being a numerate, critical thinker with tools that help make sense of data is never not my job, regardless of my job.
But let's be honest, me being adept at any of these tools is only possible because of the collective heavy lifting others have done to package and document the building blocks.
So now I get to make my contribution. Below is the complete source code, with interspersed documentation and oh-so-essential references that got me there, that I wrote to generate the data visualisation. To download a copy and use it for any purpose of your own, grab it from my GitHub account here.
COVIDBlooms¶
An animated visualisation of COVID-19 cases during the Delta variant outbreak.
Requirements¶
This is a Jupyter Notebook running a Python 3 kernel, so the most fundamental requirement is Juptyer.
Installing Jupyter typically provides the essentials: JupyterLab and Python3.
Then the extensive, specific dependencies can all be installed using pip with this one magic command (and some patience):
$ pip3 install pandas_alive geopandas descartes contextily rtree tqdm
Note that pip is not the only way to install these requirements and many prefer Anaconda. After re-visiting this dozens of times over many years, and combing the many, detailed, discussions and guides, I am very satisfied with this strategy for 2021 and MacOS:
- Avoid
condaif at all possible. - Use
brewwhenever possible (eg. for Python and Jupyter) - Then use
pip3.
I'm delighted with how well this really simple formula works today. It hasn't in the past, and may not in the future, but for now, finally and fleetingly, working with Jupyter is bliss.
Modifications¶
The dependencies, particularly pandas_alive by the very clever and generous Jack McKew, do so much of the heavy lifting. But to eek out just a bit more of what I wanted, some modifications were required. Since pip is such a convenient method for installation, I've opted to install the library as provided, and then patch it with my modifications.
All modifications can be found in my fork as three commits:
Here's one convenient way to apply each modification:
- Install
pandas_alivein the normal way withpip3 install pandas_alive. - Add
.patchto the end of the three commit urls above, and save as 610b3f4.patch, d3ddef6.patch and ddd92bb.patch. - Navigate to your installation location (which you can see in error messages, for example) and apply each patch consecutively. Eg:
$ cd /usr/local/lib/python3.9/site-packages/pandas_alive
$ patch < 610b3f4.patch
$ patch < d3ddef6.patch
$ patch < ddd92bb.patchIf you already have Jupyter open you need only Restart Python via the Kernel menu, and you're good to go.
import pandas as pd
import matplotlib.pyplot as plt
import pandas_alive
from IPython.display import HTML
import urllib.request, json
from datetime import datetime
NSW_COVID_19_CASES_BY_LOCATION_URL = "https://data.nsw.gov.au/data/api/3/action/package_show?id=aefcde60-3b0c-4bc0-9af1-6fe652944ec2"
with urllib.request.urlopen(NSW_COVID_19_CASES_BY_LOCATION_URL) as url:
data = json.loads(url.read().decode())
# Extract url to csv component
data_url = data["result"]["resources"][0]["url"]
# Read csv from data API url
df = pd.read_csv(data_url)
# Optionally, inspect data
#display(df.head())
#df[df['lga_name19'].isna()].head()
#df.describe
#df.dtypes
Clean up data to suit plotting¶
# Snip off the City/Area suffixes, (A) or (C), as well as "(NSW)" from Central Coast.
df['lga_name19'] = df['lga_name19'].str.replace(' \(.*\)$', '', regex=True)
# There's hundreds of na's. Some have postcodes but they're "weird"
# (different state or on ships?), so in general just call them "Unknown".
#df['lga_name19'].fillna("Unknown", inplace=True)
# Come to think of it, we're only interested in location so drop them altogether.
df.dropna(subset = ["lga_name19"], inplace=True)
# And while we're at it, drop correctional services for the same reason.
df.drop(df[df['lga_name19'] == 'Correctional settings'].index, inplace=True)
# Convert the date string (eg. "2020-01-25") to a datetime object.
# Bizarrely only the default ns unit works, despite only being a date string.
df['notification_date'] = pd.to_datetime(df['notification_date'])
# Group by number of records in each lga, on each day.
df_grouped = df.groupby(["notification_date", "lga_name19"]).size()
# Prepare for pandas-alive
df_cases = pd.DataFrame(df_grouped).unstack()
df_cases.columns = df_cases.columns.droplevel().astype(str)
df_cases = df_cases.fillna(0)
df_cases.index = pd.to_datetime(df_cases.index)
(Optional) Plot the default "bar chart race" animation¶
# Come alive!
#animated_html = df_cases.plot_animated(n_visible=15).get_html5_video()
#HTML(animated_html)
# Now get the geo data.
#
# This section heavily inspired by William Ye's work:
# https://medium.com/@williamye96/covid-19-in-new-south-wales-with-geopandas-and-bokeh-bb737b3b9434
import geopandas
NSW_LGA_BOUNDARIES_URL="https://data.gov.au/geoserver/nsw-local-government-areas/wfs?request=GetFeature&typeName=ckan_f6a00643_1842_48cd_9c2f_df23a3a1dc1e&outputFormat=json"
gdf = geopandas.read_file(NSW_LGA_BOUNDARIES_URL)
# Remove Lord Howe island, because it doesn't fit inside the map of NSW,
# doesn't have any cases, and the various islets occupy 62 of the 197 gdf entries!
gdf.drop(gdf[gdf['lga_pid'] == 'NSW153'].index, inplace=True)
# Remove duplicates by keeping the most recent update. Gets rid of another 6 entries.
gdf = gdf.sort_values("dt_create",ascending=False).drop_duplicates(["lga_pid"])
# Upper case the lga_name field values so they nearly match the nsw_lga__3 field values.
df["lga_name19_upper"] = df["lga_name19"].str.upper()
# And fix up the names that don't quite match
lga_replace = {"GLEN INNES SEVERN":"GLEN INNES SEVERN SHIRE", "GREATER HUME SHIRE":"GREATER HUME", "UPPER HUNTER SHIRE":"UPPER HUNTER", "WARRUMBUNGLE SHIRE":"WARRUMBUNGLE"}
df = df.replace(lga_replace)
# Re-do the grouping and prep, using the upper field instead because it's needed for the gdf merge
df_grouped = df.groupby(["notification_date", "lga_name19_upper"]).size()
df_cases = pd.DataFrame(df_grouped).unstack()
df_cases.columns = df_cases.columns.droplevel().astype(str)
df_cases = df_cases.fillna(0)
df_cases.index = pd.to_datetime(df_cases.index)
# Trim down to period of interest.
# Prior to June there's only one or two cases/day/lga. June doesn't have
# much to plot but is included because that's when lockdown started.
df_cases = df_cases['2021-06-01':]
#df_cases = df_cases['2021-10-01':] # small slice to speed up experimentation
Perform the data merge¶
# GeoSpatial charts require the transpose of that required by non-geo charts.
df_cases_t = df_cases.T
# Perform the merge of cases data with geo data, creating a new gdf with the all important "geometry" intact.
gdf_subset = gdf[["nsw_lga__3", "geometry"]]
gdf_merge = gdf_subset.merge(df_cases_t, left_on="nsw_lga__3", right_on="lga_name19_upper")
# plot_animated() can't handle any additional columns, so drop this one.
# TODO: turn it (or even better, lga_name19) into an "index" so it becomes the "geometry label"
gdf_merge.drop('nsw_lga__3', axis=1, inplace=True)
Clip the geo data to the region of interest¶
from shapely.geometry import box
b = box(150.5, -32.6, 152.2, -34.35)
box_gdf = geopandas.GeoDataFrame([1], geometry=[b], crs=gdf_merge.crs)
#fig, ax = plt.subplots(figsize=(14,14))
#gdf_merge.plot(ax=ax)
#box_gdf.boundary.plot(ax=ax, color="red")
gdf_clipped = gdf_merge.clip(b)
(Optional) Show the clipped region in context¶
#fig, ax = plt.subplots(figsize=(14,14))
#gdf_clipped.plot(ax=ax, color="purple")
#gdf_merge.boundary.plot(ax=ax)
#box_gdf.boundary.plot(ax=ax, color="red")
import contextily
fig, ax = plt.subplots(figsize=(10,10))
geo_chart = gdf_clipped.plot_animated(fig=fig,
alpha=0.35,
cmap='Oranges',
# facecolor="none", edgecolor="black",
legend=True,
vmin=0,
vmax=50,
basemap_format={'source':contextily.providers.CartoDB.Positron,
'zoom':9},
# period_label={'x':0.65, 'y':0.01, 'fontsize':36}
period_label={'x':0.65, 'y':0.01, 'fontsize':27}
)
animated_html = geo_chart.get_html5_video() # Note this is necessary whether we use `animated_html` or not...
# Optionally, play the animation now.
#HTML(animated_html)
Create the animated Bar Chart and store for later¶
# Group LGA data into regions.
# As per https://www.nsw.gov.au/covid-19/stay-safe/protecting/advice-high-risk-groups/disability/local-councils-greater-sydney
syd_lgas = ['BAYSIDE', 'BLACKTOWN', 'BLUE MOUNTAINS', 'BURWOOD', 'CAMDEN', 'CAMPBELLTOWN', 'CANADA BAY', 'CANTERBURY-BANKSTOWN', 'CENTRAL COAST', 'CUMBERLAND', 'FAIRFIELD', 'GEORGES RIVER', 'HAWKESBURY', 'HORNSBY', 'HUNTERS HILL', 'INNER WEST', 'KU-RING-GAI', 'LANE COVE', 'LIVERPOOL', 'MOSMAN', 'NORTH SYDNEY', 'NORTHERN BEACHES', 'PARRAMATTA', 'PENRITH', 'RANDWICK', 'RYDE', 'STRATHFIELD', 'SUTHERLAND SHIRE', 'SYDNEY', 'THE HILLS SHIRE', 'WAVERLEY', 'WILLOUGHBY', 'WOLLONDILLY', 'WOLLONGONG', 'WOOLLAHRA']
# Collected from sources such as https://www.dva.gov.au/sites/default/files/files/providers/Veterans%27%20Home%20Care/nsw-hunter.pdf
hunter_lgas = ['NEWCASTLE', 'LAKE MACQUARIE', 'PORT STEPHENS', 'MAITLAND', 'CESSNOCK', 'DUNGOG', 'SINGLETON', 'MUSWELLBROOK', 'UPPER HUNTER']
df_copy = pd.DataFrame(df_cases)
df_syd = df_copy[syd_lgas].sum(axis=1)
df_copy.drop(syd_lgas, axis=1, inplace=True)
df_hunter = df_copy[hunter_lgas].sum(axis=1)
df_copy.drop(hunter_lgas, axis=1, inplace=True)
df_rest = df_copy.sum(axis=1)
df_regions = pd.DataFrame({'Greater Sydney': df_syd, 'Hunter Region': df_hunter, 'Rest of NSW': df_rest})
#display(df_regions)
fig, ax = plt.subplots(figsize=(7,7))
bar_chart = df_regions.plot_animated(fig=fig, orientation='v', period_label=False, dpi=600)
animated_html = bar_chart.get_html5_video()
# Optionally, play the animation now.
#HTML(animated_html)
Create the animated Line Chart and store for later¶
fig, ax = plt.subplots(figsize=(7,7))
line_chart = df_regions.plot_animated(fig=fig, kind='line', period_label=False,
label_events={
'First case confirmed in "Bondi cluster".':datetime.strptime("16/06/2021", "%d/%m/%Y"),
'Restrictions inplace in LGAs of concern.':datetime.strptime("23/06/2021", "%d/%m/%Y"),
# 'LGAs of concern enter lockdown.':datetime.strptime("25/06/2021", "%d/%m/%Y"),
'Lockdown extended to Greater Sydney.':datetime.strptime("26/06/2021", "%d/%m/%Y"),
'Lockdown extended to all of NSW.':datetime.strptime("14/08/2021", "%d/%m/%Y"),
'Lockdown ends.':datetime.strptime("11/10/2021", "%d/%m/%Y")
}
)
animated_html = bar_chart.get_html5_video()
# Optionally, play the animation now.
#HTML(animated_html)
Finally, combine the three charts we've created into a single animation¶
from matplotlib import rcParams
rcParams.update({"figure.autolayout": False})
# make sure figures are `Figure()` instances
figs = plt.Figure(figsize=(20,11))
gs = figs.add_gridspec(2, 2, hspace=0.05, width_ratios=[10,7])
f3_ax1 = figs.add_subplot(gs[:, 0])
f3_ax1.set_title(geo_chart.title)
geo_chart.ax = f3_ax1
f3_ax2 = figs.add_subplot(gs[0, 1])
f3_ax2.set_title(bar_chart.title)
bar_chart.ax = f3_ax2
f3_ax3 = figs.add_subplot(gs[1, 1])
f3_ax3.set_title(line_chart.title)
line_chart.ax = f3_ax3
figs.suptitle(f"NSW COVID-19 Confirmed Cases Per Day, Over Time")
pandas_alive.animate_multiple_plots('multiple_charts.mp4', [geo_chart, bar_chart, line_chart], figs, enable_progress_bar=True)
Voilà!¶
Our animated chart is now available in the same folder as the Notebook file, named "multiple_charts.mp4".
Further Exploration¶
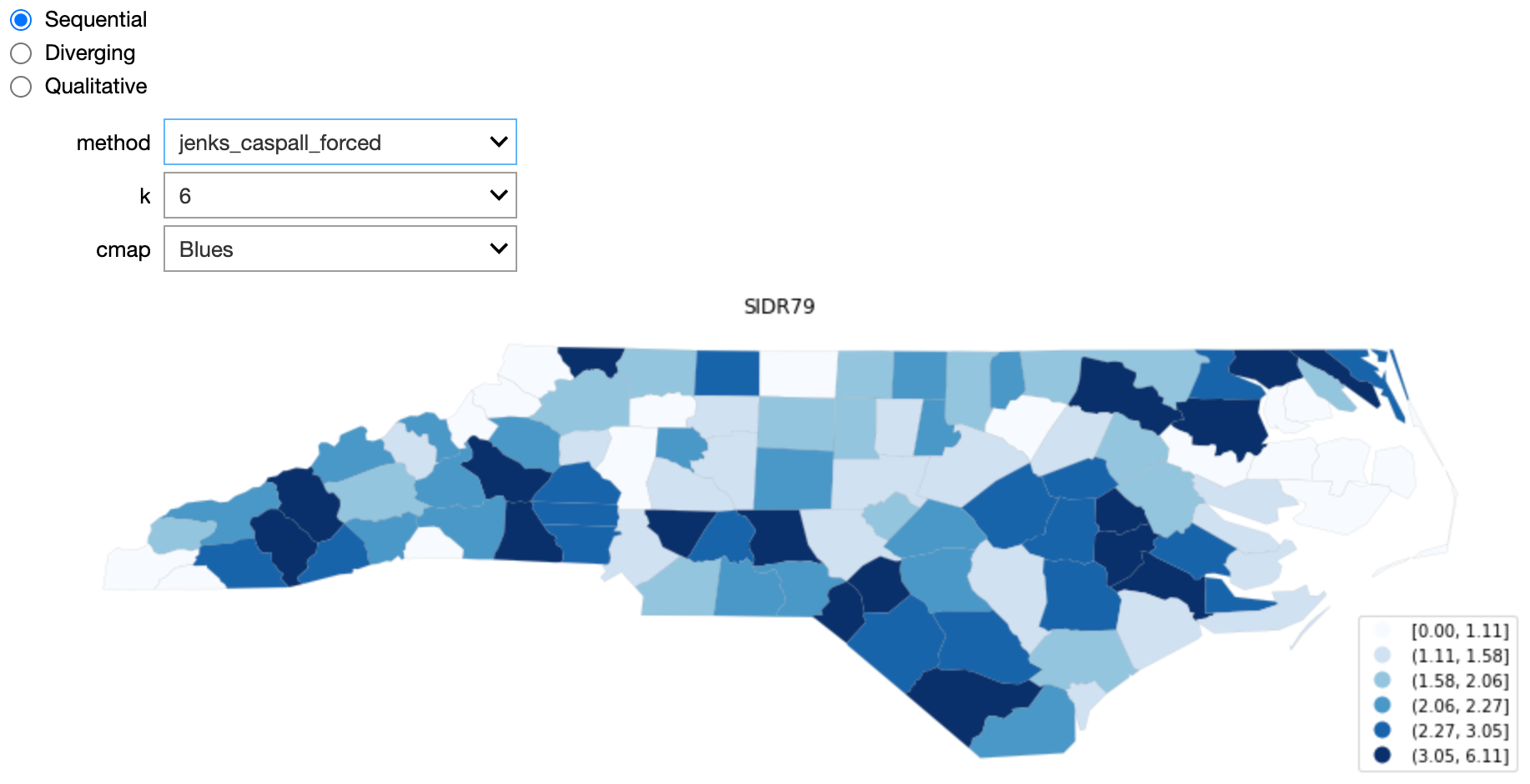
Explore the impact and usage of some of the options available when colouring choropleths.
This section is largely derived from the choropleth documentation at pysal.org.
To get started on the wormhole that is colouring choropleths, see relevant documentation from Matplotlib and GeoPandas.
Requirements¶
The additional dependencies specific to this section can all be installed using pip:
$ pip3 install palettable mapclassify libpysalimport libpysal
import mapclassify
pth = libpysal.examples.get_path('sids2.shp')
gdf_sids = geopandas.read_file(pth)
def replace_legend_items(legend, mapping):
for txt in legend.texts:
for k,v in mapping.items():
if txt.get_text() == str(k):
txt.set_text(v)
from palettable import colorbrewer
sequential = colorbrewer.COLOR_MAPS['Sequential']
diverging = colorbrewer.COLOR_MAPS['Diverging']
qualitative = colorbrewer.COLOR_MAPS['Qualitative']
from ipywidgets import interact, Dropdown, RadioButtons, IntSlider, VBox, HBox, FloatSlider, Button, Label
k_classifiers = {
'equal_interval': mapclassify.EqualInterval,
'fisher_jenks': mapclassify.FisherJenks,
'jenks_caspall': mapclassify.JenksCaspall,
'jenks_caspall_forced': mapclassify.JenksCaspallForced,
'maximum_breaks': mapclassify.MaximumBreaks,
'natural_breaks': mapclassify.NaturalBreaks,
'quantiles': mapclassify.Quantiles,
}
def k_values(ctype, cmap):
k = list(colorbrewer.COLOR_MAPS[ctype][cmap].keys())
return list(map(int, k))
def update_map(method='quantiles', k=5, cmap='Blues'):
classifier = k_classifiers[method](gdf_sids.SIDR79, k=k)
mapping = dict([(i,s) for i,s in enumerate(classifier.get_legend_classes())])
#print(classifier)
f, ax = plt.subplots(1, figsize=(16, 9))
gdf_sids.assign(cl=classifier.yb).plot(column='cl', categorical=True, \
k=k, cmap=cmap, linewidth=0.1, ax=ax, \
edgecolor='grey', legend=True, \
legend_kwds={'loc': 'lower right'})
ax.set_axis_off()
ax.set_title("SIDR79")
replace_legend_items(ax.get_legend(), mapping)
plt.show()
data_type = RadioButtons(options=['Sequential', 'Diverging', 'Qualitative'])
bindings = {'Sequential': range(3,9+1),
'Diverging': range(3,11+1),
'Qualitative': range(3,12+1)}
cmap_bindings = {'Sequential': list(sequential.keys()),
'Diverging': list(diverging.keys()),
'Qualitative': list(qualitative.keys())}
class_val = Dropdown(options=bindings[data_type.value], value=5)
cmap_val = Dropdown(options=cmap_bindings[data_type.value])
def type_change(change):
class_val.options = bindings[change['new']]
cmap_val.options = cmap_bindings[change['new']]
def cmap_change(change):
cmap=change['new']
ctype = data_type.value
k = k_values(ctype, cmap)
class_val.options = k
data_type.observe(type_change, names=['value'])
cmap_val.observe(cmap_change, names=['value'])
from ipywidgets import Output, Tab
out = Output()
t = Tab()
t.children = [out]
#t
# In this case, the interact function must be defined after the conditions stated above...
# therefore, the k now depends on the radio button
with out:
interact(update_map, method=list(k_classifiers.keys()), cmap=cmap_val, k = class_val)
display(VBox([data_type, out]))

